A Versatile Compressed Sensing Scheme For Faster And Less Phototoxic 3D Fluorescence Microscopy¶
Maxime Woringer$^1$, Xavier Darzacq, Christophe Zimmer, Mustafa Mir$^2$
- maxime.woringer@berkeley.edu
- mmir@berkeley.edu
Compressed sensing for 3D live microscopy¶
3D imaging is becoming a key technique to observe transient events in live samples. Examples of such transient phenomena include mitosis in developing fly embryos, signaling in artificial tissues in 3D matrices or in organisms.
However, such applications are often limited by one or both of the following constraints: phototoxicity or acquisition speed. Using a given setup, it is usually impossible to increase both the acquisition speed and reduce phototoxicity at a given signal-to-noise ratio (SNR).
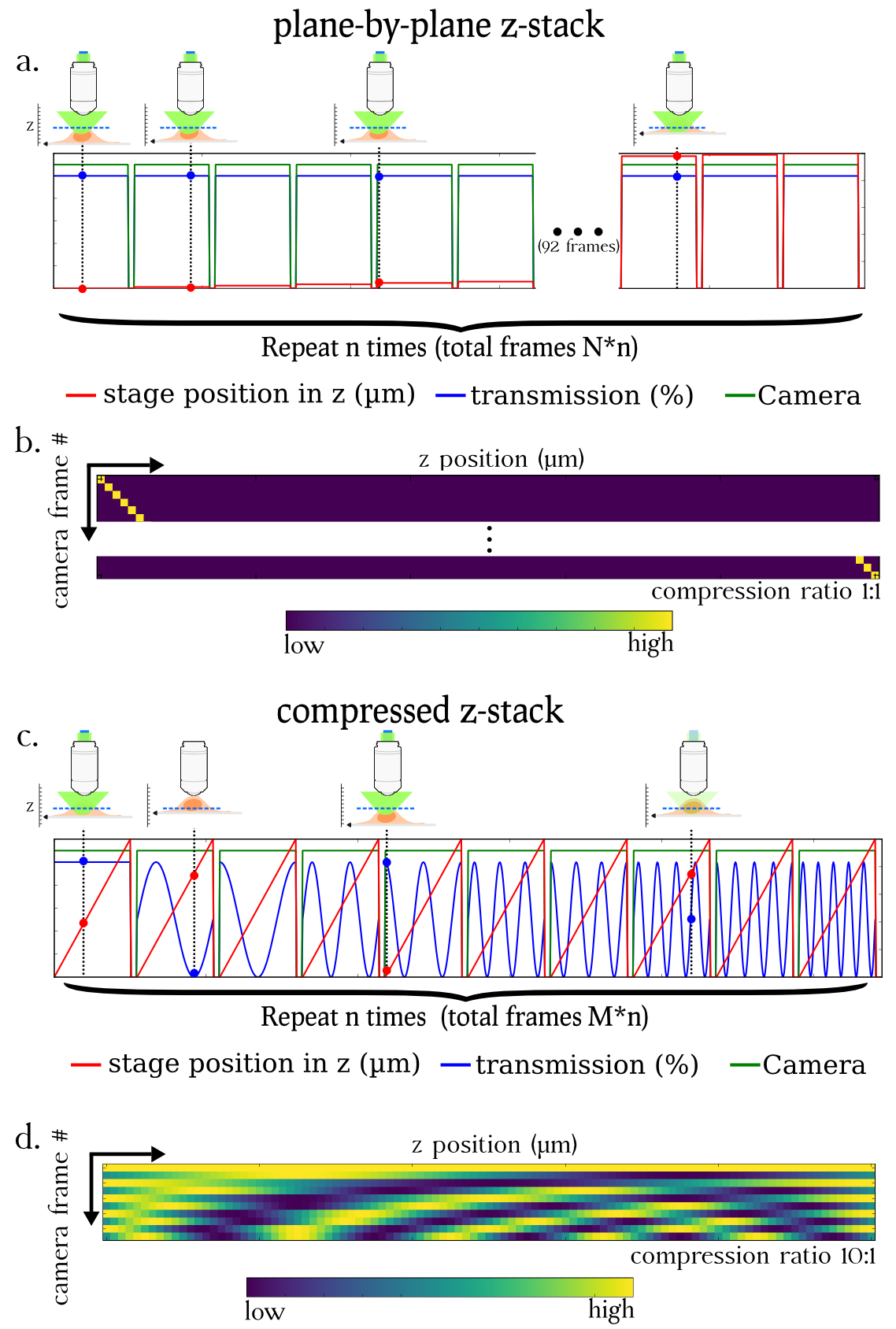
Our lab, in collaboration with the Zimmer lab in Institut Pasteur, Paris elaborated a new imaging scheme that leverages the mathematical theory of compressed sensing to perform 3D acquisitions with a reduced number of frames and with an equivalent SNR. To do so, during one camera frame, we perform an axial sweeping of the stage to successively bring into focus all the planes to image while at the same time we apply a specific light modulation. This process is repeated several times until enough information has been collected to ensure a good quality reconstruction.
Using this compressed sensing scheme, we were able to image beads and fixed mouse embryonic stem cells stained with phalloidin 5 to 10 times faster than with the traditional imaging mode. Acquisitions were performed both on a lattice light sheet microscope and a conventional epifluorescence microscope. Our scheme is very versatile and can in theory be adapted to any epifluorescence microscope with no hardware modification.
Our work is available as a preprint on BioRxiv (doi:10.1101/125815)
Data and software availability¶
The dataset used in this work is available on Zenodo, under the doi 10.5281/zenodo.439689.
The code to generate the figures is available on Github: CompressedSensingMicroscopy3D
Our port of SPIRAL-TAP is also on Github: pySPIRALTAP and doi:10.5281/zenodo.439691.
The Arduino code and the Micromanager plugin are available on Github ArduinoCompressedSensing and doi:10.5281/zenodo.495741.
Previous work¶
This work is based on preliminary work performed during my masters internship. More details about the methods and reconstructions are available on the related page
Last updated: 18 May 2017.